Abstract
Background Next-generation sequencing (NGS) is currently reforming contemporary clinical practice into precision medicine incorporating genomics-based knowledge into the clinic. Recently, multiple studies showed that mutations in epigenetic modifier genes are associated with poor treatment outcomes following tyrosine kinase inhibitor (TKI) therapy in chronic myeloid leukemia (CML), particularly after Imatinib treatment. However, it is uncertain whether they either originated from clonal hematopoiesis or the CML clone. For example, ASXL1 mutation is the most recurrent mutation at CML diagnosis, and its presence is correlated with adverse prognosis in CML. Finding of rapid reduction in ASXL1 allele frequency following TKI therapy (Kim et al, Blood 2017) raises the question of whether it is a real driver of treatment failure in CML or not. In addition, it is of interest whether 2nd generation TKI (2G-TKI) therapies can abrogate the adverse impact of somatic mutations in CML. Accordingly, the present study attempted to evaluate the prognostic impact of mutation profiles on clinical outcomes in CML patients treated either with Imatinib or 2G-TKIs.
Patients and Methods Our cohort includes a total of 394 samples including 254 samples collected at initial diagnosis and 140 samples taken during follow-up from 309 CML patients in two CML treating centers. Disease characteristics and treatment outcomes were evaluated and compared according to mutation profiles at initial diagnosis and during follow-up after TKI therapy. Treatment outcomes include any response with respect to major molecular response (MMR), molecular response (MR) with 2-log/deeper (MR2) or 4-log/deeper (MR4) with any lines of TKI therapy, failure-free survival (FFS), progression and overall survival (OS). Instead of conventional NGS, we applied smMIP-based NGS (limit of detection up to 0.2%) using our in-house CML-specific panel targeting 40 genes including epigenetic modifiers, myeloid transcription factors (TFs), tumour suppressors, genes in activated signaling, spliceosome and, cohesin complex.
ResultsPatient group and treatment summary Summary of patient characteristics are as follows: median age of 54 years; male:female n=143/111 (56.3:43.7); disease status at presentation includes chronic- (n=233; 91.7%), accelerated- (n=15; 5.9%), blastic phase (n=4; 1.6%), or N/A (n=2; 0.8%); Sokal score, low (n=61, 24.0%), intermediate (n=84, 33.1%), high (n=74, 29.1%), or N/A (n=35, 13.8%). Frontline therapy includes Imatinib (n=190; 74.8%), or 2G-TKI (n=62) including Dasatinib (n=15; 6.0%), Nilotinib (n=37; 14.7%), Bosutinib (n=10; 4.0%), or other (n=2; 0.8%).
Mutation profiles and their correlation with clinical outcomes following TKI therapy
70 mutations were detected in 57 (22.4%) out of 254 diagnostic samples, while 64 mutations were detected in 39 (27.9%) out of 140 follow-up samples.
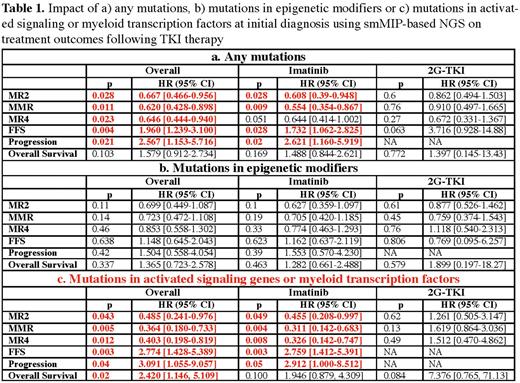
Any mutation detected at initial diagnosis is associated with worse outcomes following TKI therapy. The adverse impact of somatic mutation was mainly from myeloid TF or activated signaling pathway mutations. Patients having those mutations at initial diagnosis showed higher risks of treatment failure (HR 2.8 95% CI [1.4-5.4], p = 0.003) and progression (HR 3.1 95% CI [1.1-9.1], p = 0.04), mainly when treated with Imatinib. On the other hand, the adverse impact of epigenetic modifiers was not apparent although a trend was observed toward a lower response rate for MR2 and MMR in patients carrying mutations in epigenetic modifiers.
The adverse prognostic impact from these mutations seems to be abrogated by the use of 2G-TKI therapy, supporting the use of smMIP-based NGS for upfront TKI selection in newly diagnosed CML patients.
In the cross-sectional analysis of follow-up samples, overall mutation frequency was significantly higher in the treatment failure or progression group compared to the optimal responders (p=0.012), suggesting longitudinal tracking of mutation profiles may help identify cases of impending treatment failure or progression early.
Conclusion Our study shows that mutation profiling utilizing smMIP-based NGS can help identify patients with a potential adverse prognosis from initial diagnosis and may help upfront TKI selection. Further follow-up analysis is ongoing to evaluate the effect of TKI therapy on longitudinal kinetics of mutations in serially collected samples.
Disclosures
Žácková:Angelini Pharma: Consultancy, Speakers Bureau; Novartis: Consultancy, Speakers Bureau. Kim:Sanofi: Consultancy, Honoraria; Merck: Consultancy; Paladin: Consultancy, Honoraria, Research Funding; Pfizer: Consultancy, Honoraria, Research Funding; BMS: Research Funding; Novartis: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal